NGB
Genes track
General genes visualization
Genes are visualized from GFF/GTF/GTF3 files.
Depending on the zoom level, genes can be visualized as follows:
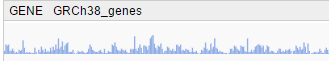
- At a low zoom level, a histogram indicating the count of features across a chromosome is shown on the whole chromosome scale.
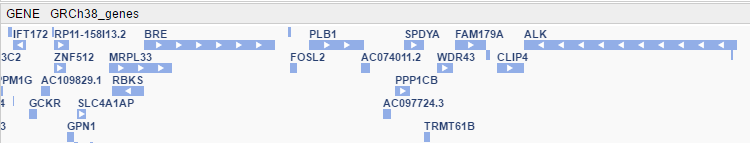
- At a medium zoom level, genes with their names and strands are shown.
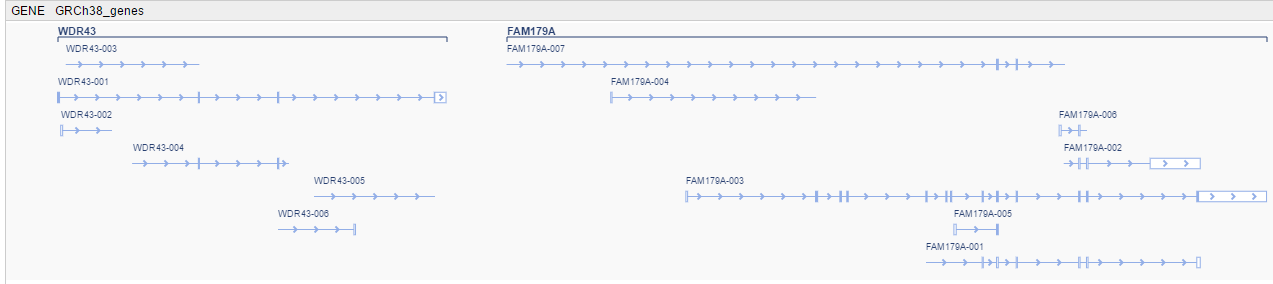
- At a high zoom level, genes with their names and their transcripts with exon-intron structure are shown.
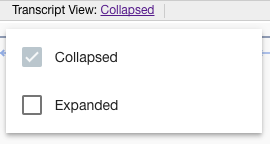
Genes track’s header contains menu of 2 items:
- Display transcripts at collapsed / expanded mode:
- “Collapsed mode” - only one canonical transcript is displayed for gene;
- “Expanded mode” - all gene’s transcripts are displayed;
- Enable / disable shortened introns mode
Shortened introns mode
Shortened introns mode is designed for cropping regions with introns (taking gene’s introns from particular track) for all tracks. At this mode:
- Zooming in / out is disabled
- Moving to a position is disabled
- Ruler’s local track displays shortened regions (as orange bars), as well as total and actual (in brackets) regions sizes

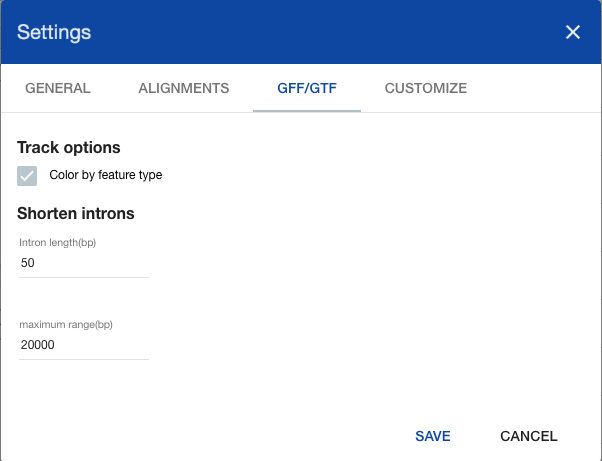
Shortened introns mode has 2 configurable parameters:
- Introns length - the size in base pairs that is not cropped from intron’s borders. Such offsets are displayed as semi transparent bars at local ruler track:

- Maximum range (bp) - the maximum range at which shortened introns mode is available. If current selected range is larger then that value a message is displayed at genes track’s header
These parameters are managed at GFF/GTF section of global settings window